Research
The objective of our research program is to study the mechanisms of resistance to antibacterials (antibiotics, metals, phages) and to understand how phages control pathogenic bacteria in nature.
Why?
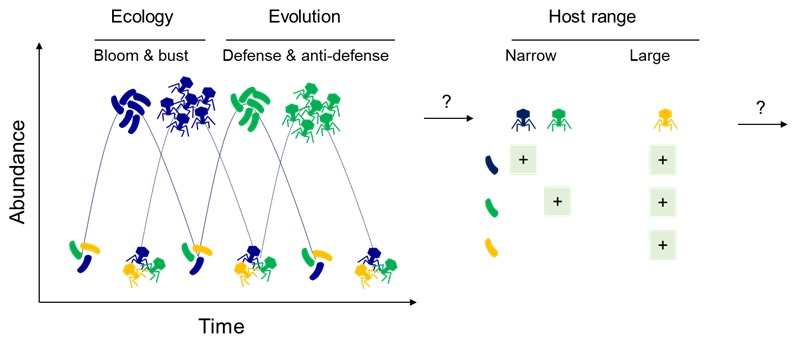
By 2050, multi-antibiotic-resistant bacteria could cause 10 million deaths per year. Environmental degradation (pollution, decline in diversity) plays a key role in the development and spread of antibiotic resistance. But the environment is also a source for discovering therapeutic solutions that are more respectful of nature. For example, bacteriophages (or phages) are natural predators of bacteria and are very abundant in aquatic environments, like sewers near hospitals or aquaculture farms. Phages multiply in their host and are very specific to certain bacteria. We will look for phages that only infect pathogens and do not infect innocuous or useful bacteria. It is sometimes necessary to use a mix of different phages (called a cocktail) to kill all of the pathogens, either because the phage only infects part of the pathogenic bacteria, or because the bacteria develop resistance, just as with antibiotics. However, although the appearance of resistance to phages is often observed in the laboratory, it seems that in nature, things are different. In a game of “arms race”, the bacteria develops mechanisms to resist the phages, which counterattack by developing counter-resistance. As the joint evolution of the phage and the bacteria (or coevolution) can be to the detriment of other capacities, it is said to have a cost. For example, it can result in a reduction in the virulence of the bacteria, a major advantage in the treatment of an infection.
How?
We will study the mechanisms and evolution of the interaction between bacteria and their phages in natural populations. We will use marine bacteria from the Vibrionaceae (or vibrios) family as a model system. Vibrios include human pathogens responsible for gastroenteritis in Canada (e.g. V. parahaemolyticus) and animal pathogens (fish, crustaceans and shellfish). Our study system therefore promotes translational research within the framework of a priority research area for UdeM “one health”. Based on a large collection of isolates, we will explore the mechanisms involved in the coevolution of phages and bacteria, how this coevolution affects phage specificity and bacterial pathogenicity. The proposed work is innovative because it combines ecology, population, comparative and functional genomics as well as microscopy to explore in “real time” the behavior and evolution of these microbes.
Research Projects from the members of the lab

Manon Lang (Post-Doc)
We have unveiled a new family of satellites known as Phage-Inducible Chromosomal Minimalist Islands (PICMIs) (Rubén Barcia-Cruz et al., 2024). These satellites are characterized by reduced gene content and lack genes for capsid remodeling, instead packaging their DNA as a concatemer. PICMIs rely on virulent phages to disseminate to other bacteria and shield their hosts from competitive phages without impeding their helper phage.
Manon will undertake the characterization of the molecular mechanisms governing each stage of the PICMI lifestyle. This includes elucidating activation by the helper phage, excision from the host genome, replication via rolling circle, packaging into the capsid, and examining its role in bacterial host resistance. Additionally, she will delve into the structure-function relationship of various putative regulator AlpA proteins identified across a broad spectrum of satellites.

Carine Diarra (Lab manager/technician)
Carine is tasked with managing various aspects of laboratory operations, including ordering supplies, maintaining stock inventor and coordinating the collection of microorganisms. In addition to her administrative duties, she actively participates in conducting large-scale experiments to advance specific research projects.
Presently, Carine collaborates with Jeff, our summer trainee, on investigating the infection dynamics of phages belonging to the Schizotequatrovirus family. Their research focuses on examining infection patterns at both population and single-cell levels, employing classical virology techniques alongside timelapse microscopy and cytometry methods. This study is facilitated by the resources provided by the CIB’s microscopy and cytometry platform.

Martin Lamarche (Scientific Advisor)
Martin is tasked with overseeing the development of molecular biology approaches for phages and vibrios, including staff training. We employ allelic replacement techniques to manipulate bacterial genes linked to specific phenotypes. In our investigation of phage genes related to infection, we will utilize genetic tools such as homologous recombination and CRISPR-based counter-selection systems, collaborating with Damien Piel from Alexander Harm’s lab at ETH Zürich. Martin manages our droplet digital PCR machine (Biorad QX600) and customizes protocols to suit our specific research questions and model systems.
Currently, he collaborates with Charles Bernard from Eduardo Rocha’s lab at the Pasteur Institute, focusing on characterizing defense and antidefense elements involved in vibrio-phage coevolution.

Dario Cueva (PhD Student)
From our published work (Piel et al., 2022) we now possess a clear understanding of the specific genetic components that warrant our attention for monitoring co-evolving lineages in ‘real time’ in the environment. Specifically, we have identified clades nestled within V. crassostreae and species of phages that demonstrate concordant adsorption. We also showed that coevolution depends on bacterial defenses and phage counter defenses. Armed with this knowledge, we scaled up our sampling on a time-series sampling initiative within an oyster farm to test our hypothesisthat predation by virulent phages affects V. crassostreae infection dynamics for two reasons: it reduces pathogen density and it selects for less virulent, phage-resistant strains of vibrios.
Dario’s project is an integral part of the study on the eco-evolutionary dynamics of phages in nature. Employing ddPCR, Dario will systematically monitor the abundance of each vibrio clade and phage species over time and space to formally demonstrate that the bloom of predator coincides with the decline of the prey. Focusing on the V1 clade, Dario will explore the origin and consequences of the small genome size of these strains.

Jeff Liang (Post-Doc)
Jeff will work on bioinformatic analysis with the data collections built up from years of inquiry by the Le Roux group into this phage-vibrio system. His research will focus on investigating genetic underpinnings of the strain-wise differences in pathology and ecology in related strains of V. crassostreae. Species in the genus Vibrio often exhibit an open pangenome and high levels of horizontal gene transfer, features which are likely to contribute to species’ adaptations to their ecological niches.
While the clustering of these strains by core genome alignments is known to be inter-related to virulence and phage susceptibility, the contribution of diversity across the accessory genome is not known. Jeff’s project will make inquiries into the evolutionary dynamics of the stratified oyster-Vibrio-phage system through comparative population genetic and reverse ecological approaches. This will include assessments of historical selection pressure, inferences of population history patterns, and phylogeographical assessments of recombinant populations. These studies will be conducted in partnership with the Shapiro group at McGill University and the Rocha group at the Pasteur Institute.
