Research
The objective of our research program is to study the mechanisms of resistance to antibacterials (antibiotics, metals, phages) and to understand how phages control pathogenic bacteria in nature.
Why?
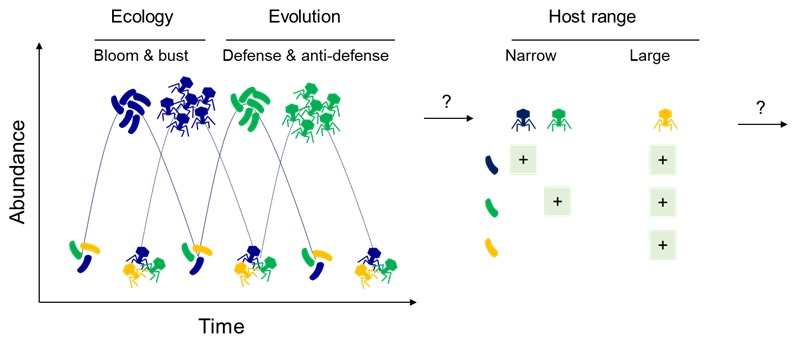
By 2050, multi-antibiotic-resistant bacteria could cause 10 million deaths per year. Environmental degradation (pollution, decline in diversity) plays a key role in the development and spread of antibiotic resistance. But the environment is also a source for discovering therapeutic solutions that are more respectful of nature. For example, bacteriophages (or phages) are natural predators of bacteria and are very abundant in aquatic environments, like sewers near hospitals or aquaculture farms. Phages multiply in their host and are very specific to certain bacteria. We will look for phages that only infect pathogens and do not infect innocuous or useful bacteria. It is sometimes necessary to use a mix of different phages (called a cocktail) to kill all of the pathogens, either because the phage only infects part of the pathogenic bacteria, or because the bacteria develop resistance, just as with antibiotics. However, although the appearance of resistance to phages is often observed in the laboratory, it seems that in nature, things are different. In a game of “arms race”, the bacteria develops mechanisms to resist the phages, which counterattack by developing counter-resistance. As the joint evolution of the phage and the bacteria (or coevolution) can be to the detriment of other capacities, it is said to have a cost. For example, it can result in a reduction in the virulence of the bacteria, a major advantage in the treatment of an infection.
How?
We will study the mechanisms and evolution of the interaction between bacteria and their phages in natural populations. We will use marine bacteria from the Vibrionaceae (or vibrios) family as a model system. Vibrios include human pathogens responsible for gastroenteritis in Canada (e.g. V. parahaemolyticus) and animal pathogens (fish, crustaceans and shellfish). Our study system therefore promotes translational research within the framework of a priority research area for UdeM “one health”. Based on a large collection of isolates, we will explore the mechanisms involved in the coevolution of phages and bacteria, how this coevolution affects phage specificity and bacterial pathogenicity. The proposed work is innovative because it combines ecology, population, comparative and functional genomics as well as microscopy to explore in “real time” the behavior and evolution of these microbes.
Research Projects from the members of the lab

Coevolution of PICMI satellites and their helper phages
Postdoc: Manon Lang (Apr 2024-)
Poster presentation at CSM Montréal, June 2025
Phage satellites are MGEs that hijack phage machinery for their own dissemination. We previously identified a distinct family, PICMIs (Phage-Inducible Chromosomal Minimalist Islands), broadly distributed in marine Vibrionaceae (see Rubén Barcia-Cruz et al., 2024). PICMIs have reduced gene content, encode no capsid remodeling functions, package DNA as concatemers, and integrate near fis. Three core proteins (Int, AlpA, Prim) ensure excision and replication, but encapsidation mechanisms remain unknown.
PICMIs depend on virulent phages for dissemination, protecting hosts from competitor phages without noticeably impairing their helper. In fact, only ~10% of our prototype helper is parasitized, supporting the hypothesis. Using a ΔPICMI strain and viromes from 2021 sampling, we isolated new “bad helpers” that could not trigger excision or early gene expression and thus produced no parasitized capsids. We hypothesized that highly parasitized helpers (>50%) would not be easily isolated by plaque assays, similar to the V. cholerae PLE, which completely abolishes ICP1 production.
To test this hypothesis and search for better helper, we engineered PICMI with resistance, replication (R6K), and transfer (RP4) cassettes, enabling plasmid production in E. coli, transfer and insertion into Vibrio strains (V. chagasii and V. crassostreae) susceptible to at least one phage. This strategy identified ~15 phages affected by PICMI, with >50% capsid parasitism. This higher parasitism is not linked to more efficient excision or replication, but to improved encapsidation. With these “good helpers,” we revisited PICMI gene functions and found the gene that is essential for encapsidation.
These results redefine phage–satellite interactions: helpers can evade PICMI either by blocking early gene activation or by preventing encapsidation

Carine Diarra (Lab manager/technician)
Carine is tasked with managing various aspects of laboratory operations, including ordering supplies, maintaining stock inventor and coordinating the collection of microorganisms. In addition to her administrative duties, she actively participates in conducting large-scale experiments to advance specific research projects.
Presently, Carine collaborates with Jeff, our summer trainee, on investigating the infection dynamics of phages belonging to the Schizotequatrovirus family. Their research focuses on examining infection patterns at both population and single-cell levels, employing classical virology techniques alongside timelapse microscopy and cytometry methods. This study is facilitated by the resources provided by the CIB’s microscopy and cytometry platform.

Martin Lamarche (Scientific Advisor)
Martin is tasked with overseeing the development of molecular biology approaches for phages and vibrios, including staff training. We employ allelic replacement techniques to manipulate bacterial genes linked to specific phenotypes. In our investigation of phage genes related to infection, we will utilize genetic tools such as homologous recombination and CRISPR-based counter-selection systems, collaborating with Damien Piel from Alexander Harm’s lab at ETH Zürich. Martin manages our droplet digital PCR machine (Biorad QX600) and customizes protocols to suit our specific research questions and model systems.
Currently, he collaborates with Charles Bernard from Eduardo Rocha’s lab at the Pasteur Institute, focusing on characterizing defense and antidefense elements involved in vibrio-phage coevolution.

Defense island and the role of BREX in plasmid exclusion (Clade V1)
PhD student: Dario Fernando Cueva (Sept 2024- )
Poster presentation at CSM Montréal, June 2025
From our published work (Piel et al., 2022) we now possess a clear understanding of the specific genetic components that warrant our attention for monitoring co-evolving lineages in ‘real time’ in the environment. Specifically, we have identified clades nestled within V. crassostreae and species of phages that demonstrate concordant adsorption. We also showed that coevolution depends on bacterial defenses and phage counter defenses. Armed with this knowledge, we scaled up our sampling on a time-series sampling initiative within an oyster farm to test our hypothesis that predation by virulent phages affects V. crassostreae infection dynamics for two reasons: it reduces pathogen density and it selects for less virulent, phage-resistant strains of vibrios.
Dario’s project is an integral part of the study on the eco-evolutionary dynamics of phages in nature. Employing ddPCR, Dario systematically monitored the abundance of each vibrio clade and phage species over time and space. He found that despite strong fluctuations in bacterial and phage abundances, no simple predator–prey cycles were observed. Instead, host resistance, spatial refuges, multi-phage predation, and lysogeny appear to promote long-term coexistence (see our preprint https://www.biorxiv.org/cgi/content/short/2025.10.10.681744v1).
Dario is now exploring the origin and consequences of the small genome size of these strains. Vibrio clade V1 isolates have reduced genomes (~5.1 Mb) and strikingly zero plasmid for most strains. Conjugation assays showed that many strains are unable to acquire or maintain plasmids. This non-permissiveness correlates with the presence of a genomic island encoding multiple defense systems. Deletion of the island restores plasmid uptake with a three-log increase in conjugation frequency. Gene-by-gene knockouts are underway to identify causal genes and validate by complementation. Further work will decipher the mechanism of activity of this anti-plasmid element.

Unexpected stability of phages and diversity of MGEs in V. crassostreae
Postdoc: Jeff Liang (Aug 2024- )
Collaboration: Eduardo Rocha
Preprint : https://www.biorxiv.org/cgi/content/short/2025.10.10.681744v1
Poster presentation at CSM Montréal, June 2025
Phages are generally considered fast-evolving entities, driven by mutation and recombination. We tested this hypothesis in a natural intertidal environment, an oyster farm on the Brittany coast (France), through longitudinal sampling that yielded over 1,000 lytic phages and 600 Vibrio crassostreae isolates. Surprisingly, some phage and bacterial lineages proved highly persistent, with nearly identical virulent phages recovered after four years at the same location. The infection network remained modular, consistently involving the same V. crassostreae clades (closely-related clusters of strains with shared habitats and virulence features) and phage genera. This stability may be explained by winter persistence of V. crassostreae in wild oysters, seasonal active proliferation in summer, and limited phage degradation in the environment. Despite strong fluctuations in bacterial and phage abundances, no simple predator–prey cycles were observed. Instead, host resistance, spatial refuges, multi-phage predation, and lysogeny appear to promote long-term coexistence.
Oysters also emerge as hotspots for the acquisition and selection of mobile genetic elements (MGEs). Oyster-associated isolates (clades V2–V5, V8) harbor more plasmids, prophages, and phage-satellites, and display significantly larger genomes compared to water-column isolates (clade V1). Plasmids are enriched in colonization traits (metal resistance, antimicrobial peptides, virulence factors), while prophages and satellites show a higher density of anti-phage defense systems. Virulent phages are abundant in oysters, suggesting strong selective pressure. Notably, oyster isolates revealed unexpected MGE diversity, including extrachromosomal circular or linear phage–plasmids and satellite–plasmids, opening new research avenues. These findings highlight how ecological constraints shape viral lineage stability: vibrios evade oyster immunity, restrict phage diversity through receptors and defense systems, while satellites exploit their helper phages.

Schizotequatroviruses: viral viperin and phage–host coexistence
MSc student: Caleb Veilleux Gravel (Jan 2025-2027)
This MSc project extends the study of Schizotequatroviruses (see our paper in ISME J. https://doi.org/10.1093/ismejo/wraf063) by focusing on two original aspects: (1) the presence of a phage-encoded viperin gene, and (2) atypical infection cycles allowing coexistence with hosts.
First, we are investigating the function of viperin homologs—typically antiviral proteins in both prokaryotes and eukaryotes, here carried by phages. Viperins convert CTP into ddhCTP, a modified nucleotide that blocks viral RNA polymerases. Three phage viperins and five host-derived homologs were cloned under a pBAD promoter in E. coli K-12, and their anti-phage activity is being tested against ~30 phages from the Basel collection (collab. Damien Piel & Alexander Harms). Activity is also evaluated in a T7 polymerase/GFP reporter strain.
Second, we are characterizing atypical infection cycles. In some hosts, phages persist without rapid lysis: part of the population produces phages continuously while the majority become resistant. Fluorescently labeled phages (Neon Green or PHOLLOW, with Martin Lamarche) will be tracked by time-lapse microscopy and flow cytometry (with Carine Diarra & Yves Brun). FISH experiments will visualize viral DNA to clarify infection strategies: pseudolysogeny, lysis inhibition, or persistent infections.
This work will form the basis for Caleb’s PhD project in the lab. In parallel, with José Penadés and da Costa labs (Imperial College, London), cryo-EM is being used to determine Schizotequatrovirus structure.

Discovery of a new extrachromosomal phage satellite
MSc student: Béatrice Lefèvre (Jan 2025-2027)
We discovered a novel extrachromosomal element in Vibrio, named cf-PISP (capsid-forming Phage-Inducible Satellite–Plasmid) (see
https://www.biorxiv.org/cgi/content/short/2025.10.10.681744v1). Prototype TG54 is a 15.8 kb circular element carrying plasmid-like maintenance genes (parA, repA, ProQ/FinO-like), consistent with episomal replication (validated by PCR). TG54 also encodes structural proteins for capsid assembly (capsid, portal, protease, adapter, closure) and an stl-like regulator. The absence of integrase and capsid sequence related to Cf-PICI rather than phage, confirm its unique status: neither phage nor PICI, but an episomal satellite–plasmid. Related elements were found in other Vibrio strains, but not in other genera. We hypothesize that, unlike chromosome-integrated satellites (PICI, CfPICI, P4, PLE, PICMI), Cf-PISP combines vertical plasmid transmission with horizontal spread via hijacked phage capsids. Béatrice’s project will explore 1) the plasmid-like replication of Cf-PISP; 2) the packaging of the element by its own capsid; 3) its transmission.
This project establishes cf-PISP as a novel phage–satellite interaction model, broadening known MGE diversity and providing an excellent MSc thesis. It will benefit from collaborations with Rocha lab (Pasteur Institute) and Jose Penades lab (Imperial College, London).

haracterization of the phage-plasmid Kali
Postdoc: Léa Verena Zinsli (Feb 2025- )
Among 562 Caudoviricetes prophages identified in V. crassostreae isolates, 37 (6.6%) were extrachromosomal. One, a ~35 kb siphovirus-like phage-plasmid named Kali (LG49/TG34), is present in 5/145 genomes from clade V5 (approx. three copies/genome). One strain has since lost Kali, suggesting instability.
Kali is intriguing because it:
- Is active in oyster hemolymph
- Shows diverse infection outcomes in a 112 × 73 infection matrix: clear plaques in some V5 subgroups (lytic behavior), turbid phenotypes (adsorption without viral production) in others, or full resistance.
Clarifying lysogeny vs. defense phenotypes is essential for GWAS studies on resistance and latency/lysis determinants (collab. Charles Bernard & Eduardo Rocha). As first step toward redefining the future of the infections (lytic, lysogen, restricted or abortive) we (i) cured Kali from native strains, (ii) inserted a CmR marker into lysogenic vs. lytic variants, (iii) induced with mitomycin and tracked particle production by Southern blot, ddPCR, and plaque assays.
Strains carrying Kali often host multiple prophages, opening future work on polysogeny regulation.
